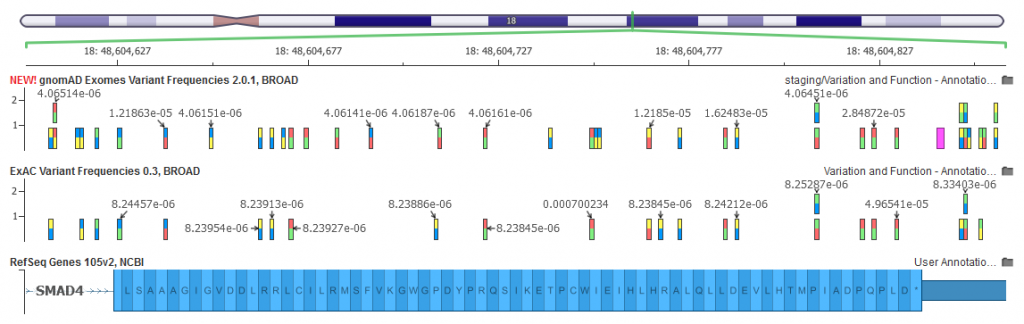
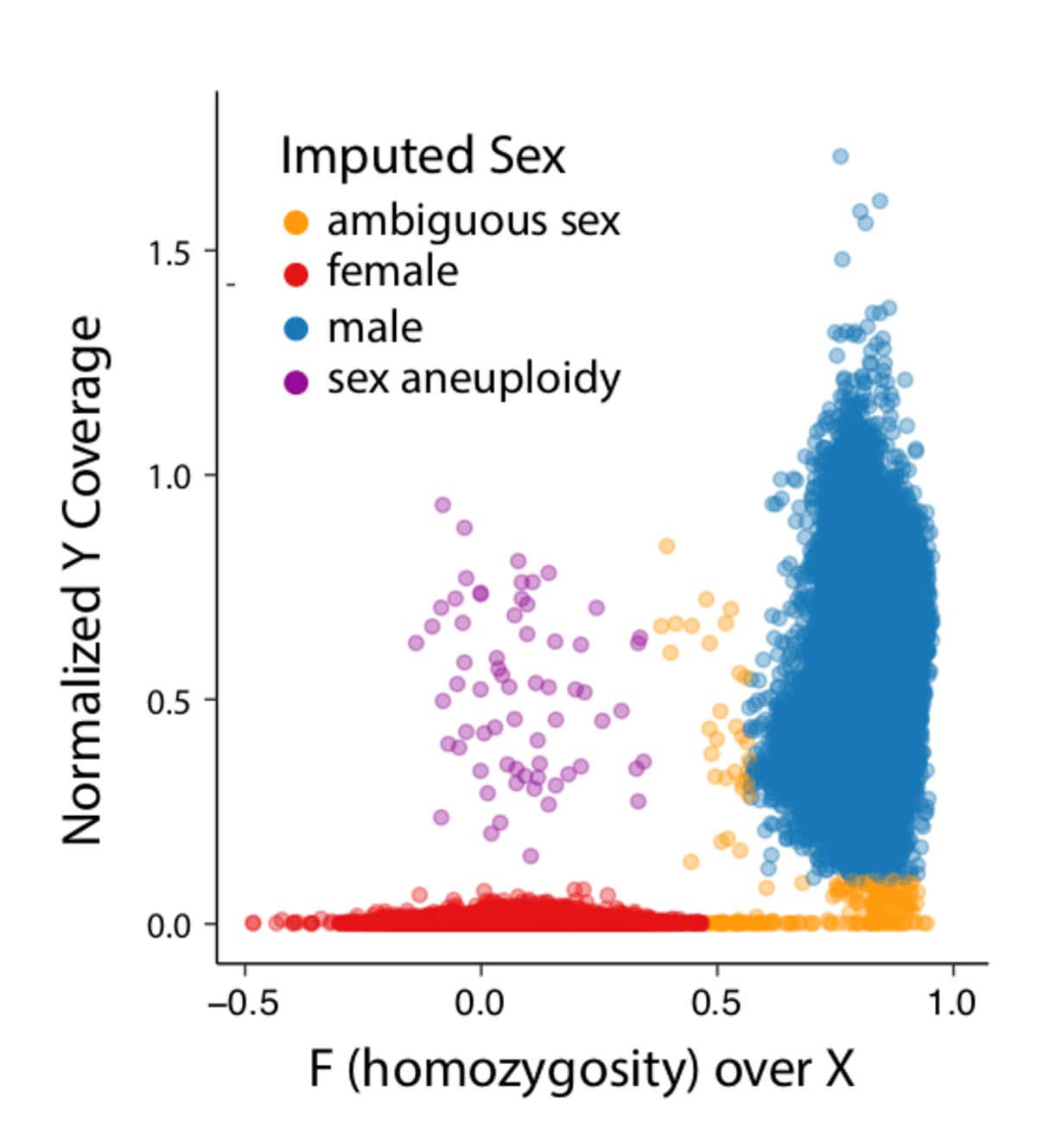
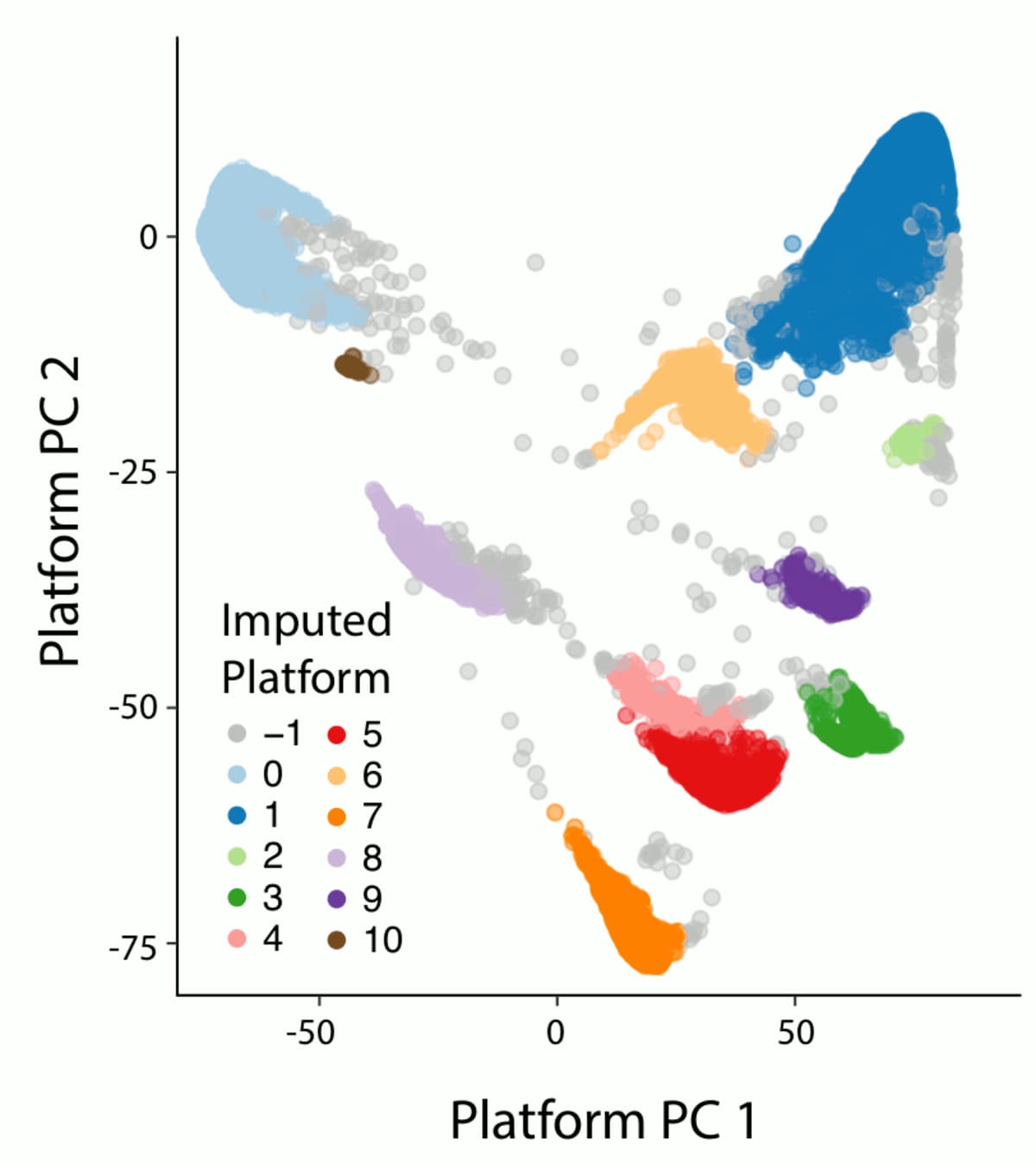
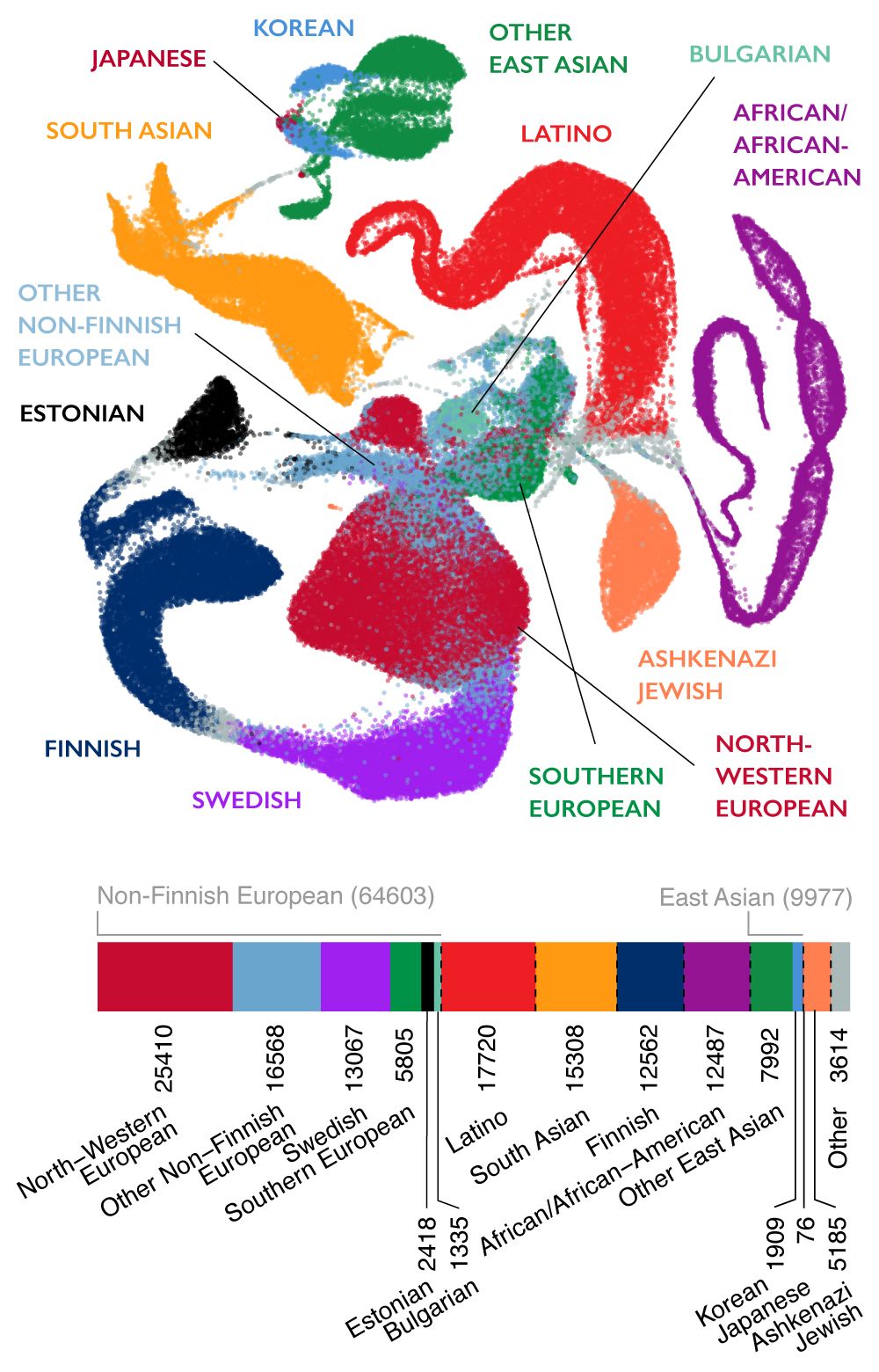
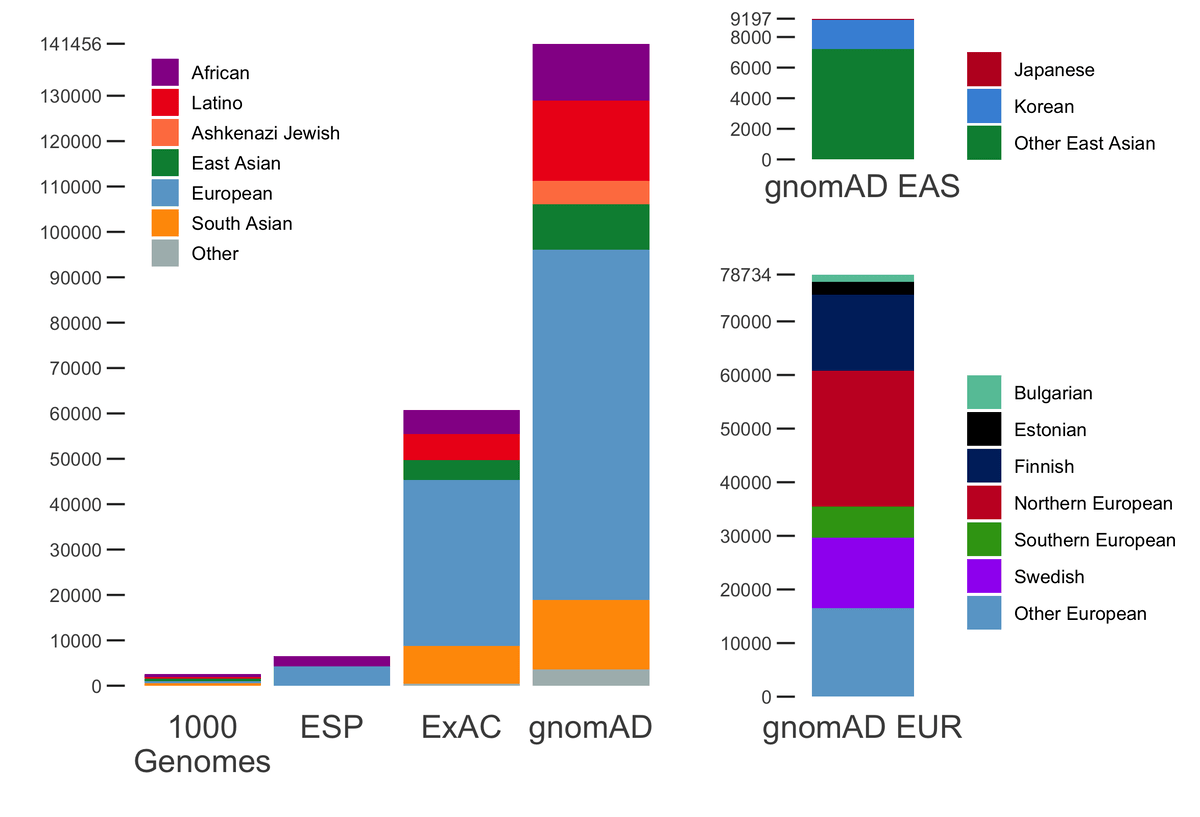
All of the variants absent from gnomAD were rare in the EHVol cohort... | Download Scientific Diagram

The Genome Aggregation Database (gnomAD) @nature @Jaivikshastram - Jaivikshastram Education & Research Foundation

Womens Gnomad Tshirt Funny Nomad Traveller Gnome Fairytale Graphic Tee (Light Heather Grey) - 3XL Womens Graphic Tees - Walmart.com

Genome Aggregation Database (gnomAD): Now available on Azure Open Datasets - Microsoft Community Hub

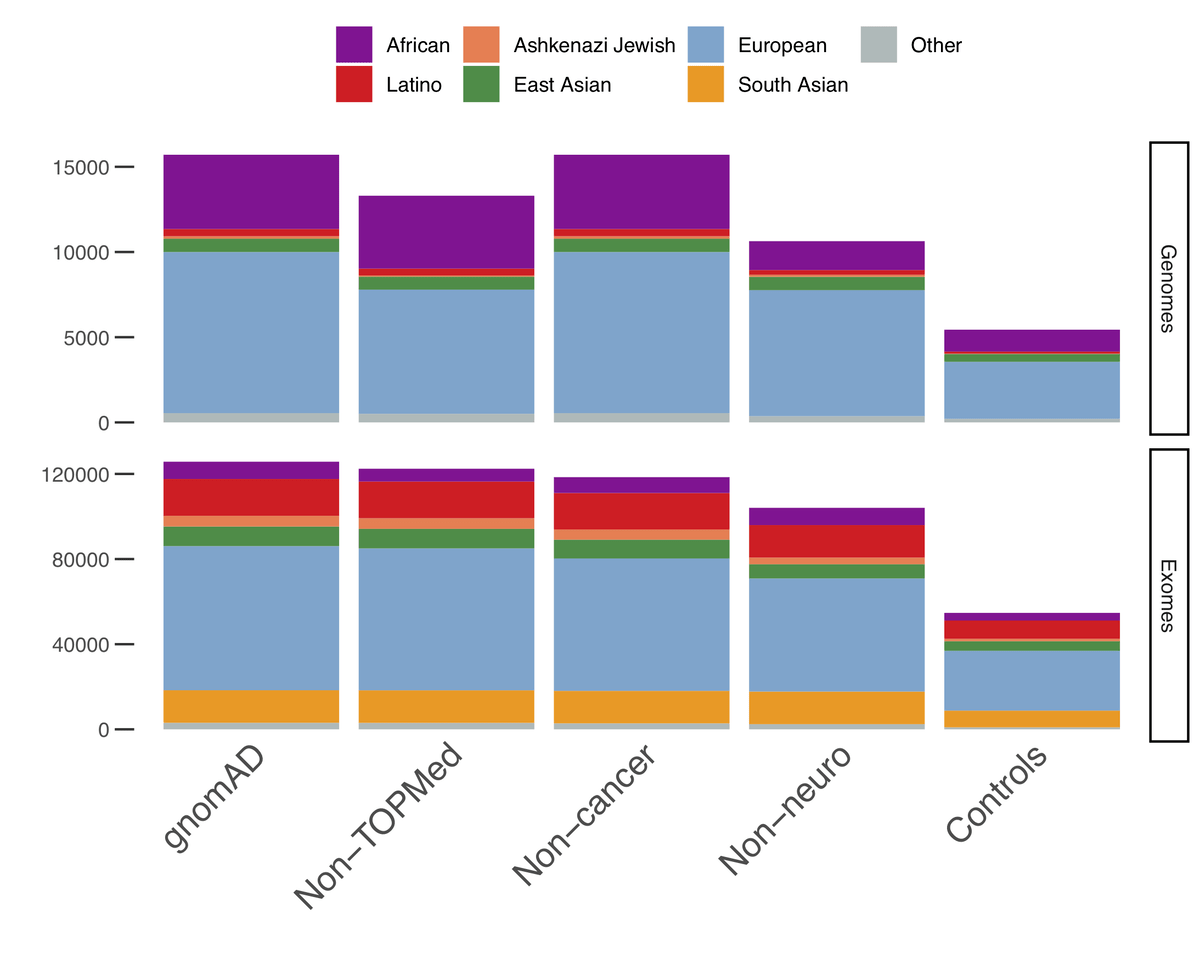
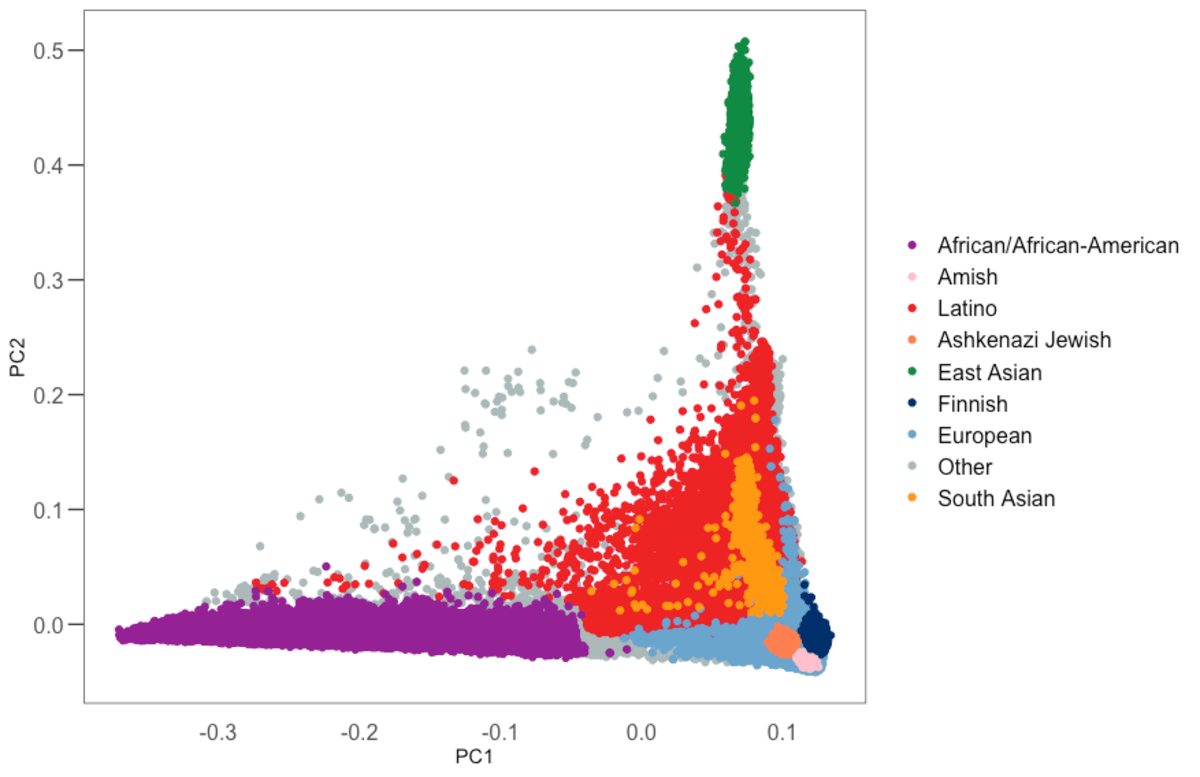
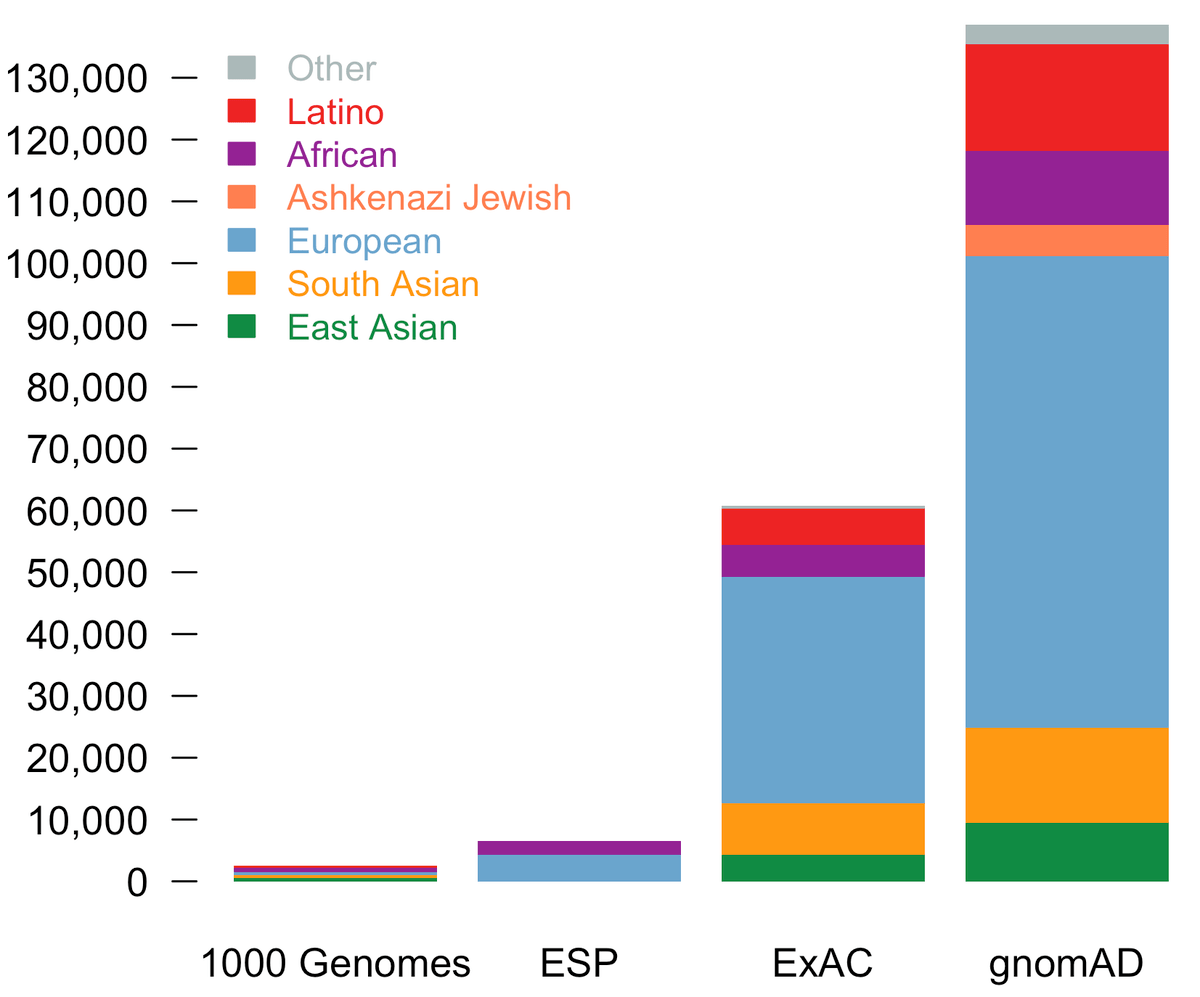
Populations represented in the gnomAD database. An example of various... | Download Scientific Diagram